The variety of instrumentation allows for capabilities ranging from single cell RNA-seq or epigenomic or multiomic analyses, spatial transcriptomic analysis of both frozen and FFPE tissue, whole genome, exome, epigenome and transcriptome sequencing as well as a focused analysis of the genotypes and DNA methylation using array technology. Services are available for a variety of study designs extending from a few laboratory samples to large clinical projects involving hundreds or thousands of samples. The full bioinformatics service is also available for data analysis.
The facility receives either tissue/cells, DNA/RNA or customer-generated NGS libraries. Samples are processed based on agreement reached during consultations on the design of the experiment. Well-experienced staff members are available for assisting in project operations. The facility develops new applications to accommodate state-of-the-art NGS technologies. Additionally, data analysis service includes sequence-read alignment, secondary analysis (quantitation, variant calling, functional annotation, visualization, etc.) and follow-up with the interpretation of the results. The facility also provides support for grant writing and hands-on training for students and postdocs in NGS processing. All projects, submissions and pricing checks must be done through iLab.
The facility resides in 5,000 square feet of newly renovated space, encompassing separate “pre-amplification” and “post-amplification” rooms to prevent any contamination of PCR-amplified materials to pre-processed input DNA/RNA samples.
For general inquiries, email GenomeSciences@pennstatehealth.psu.edu.
Jump to topic
Search
Instrumentation and Services
A variety of instrumentation is available in the Genome Sciences Facility for sample processing, sample quantitation and quality control, qPCR/digital PCR, microarray analyses, next-generation sequencing and other needs.
Single cell analysis
Next-Generation Sequencing (NGS)
Microarray Analyses
qPCR/Digital PCR
Sample Processing
- QIAsymphony DNA/RNA Extraction Robot
- Bullet Blender (high-throughput tissue homogenizer)
- Covaris Adaptive Fcused Acoustics Ultrasonicator E-Series
Sample Quantitation and Quality Control
- Agilent 2100 Bioanalyzer
- Nanodrop ND-1000 Spectrophotometer (available for use from 10 a.m. to 3 p.m. weekdays in Room C3747)
- Qubit Fluorometer
Software
- Illumina GenomeStudio
- Ingenuity Pathway Analysis (email the IS Service Desk at help@pennstatehealth.psu.edu or call 1-833-577-HELP (4357) for details)
Nucleic Acid Isolation
Nucleic acid isolation is available using the QIAsymphony DNA/RNA Extraction Robot.
The QIAsymphony SP enables sample preparation of DNA, RNA, bacterial and viral nucleic acids from whole blood, saliva and buffy coat among other sample types.
Available Preps
- 200 ul Whole Blood DNA prep
- 400 ul Whole Blood DNA prep
- 1000 ul Whole Blood DNA prep
- 200 ul DNA prep buffy coat
- 400 ul DNA prep buffy coat
- 400 ul RNA prep
- 800 ul RNA prep
- PAXgene_RNA
- 1000 ul Saliva DNA prep
Covaris Adaptive Focused Acoustics Ultrasonicator
This equipment, providing DNA and chromatin shearing capabilities, is available in room C2706. The Covaris enables shearing of samples without thermal damage. The equipment is available on a sign-up basis.
Other options for sample extraction of RNA/DNA may be found in iLab.
Genome Sciences has Illumina sequencing instrumentation – MiSeq and NovaSeq6000 – for both focused and large-scale sequencing by synthesis. Specific library construction services are available. Investigators who have a sequencing project should initiate the project through iLab. to receive a project ID before submitting samples.
MiSeq
For library construction, the facility recommends Illumina library preparation kits, and most other library preparation kits with Illumina adaptors will work with the MiSeq.
Illumina offers a tool to help investigators choose the appropriate library prep and array kit.
NovaSeq 6000
This offers scalable throughput and flexibility for virtually any sequencing method, genome and scale of project.
Information on all of these options may be found in iLab; contact Genome Sciences at genomesciences@pennstatehealth.psu.edu if necessary.
The Genome Sciences Facility provides a variety of library preparation services including whole genome, whole exome, ChIP-seq, Methylation-seq, RNA-seq and targeted resequencing. Listed below are currently operated services, but the facility is expanding the service to meet the needs of each investigator.
- High throughput single-cell RNA-seq/ATAC-seq/multiome using 10x Genomics platform
- 10x Genomics visium spatial transcriptomics
- Whole-genome sequencing library prep
- Whole-exome sequencing (Human) (minimum input DNA 1 ng for intact DNA, under R and D for FFPE DNA)
- Exome (other species, mouse, bovine, zebrafish)
- ChIP-sequencing library prep (minimum input DNA 0.5 ng)
- Low-input ChIP-seq library prep (minimum input DNA 0.05 ng)
- Methylation sequencing -ERRBS (enhanced reduced representation of bisulfite sequencing) library prep
- PolyA RNA seq library prep (strand-specific, minimum input RNA 50 ng)
- Total RNA seq library prep (rRNA depleted, human, mouse, rat, standard rRNA-depletion, strand-specific, minimum input RNA 100 ng)
- Low-input RNA-seq library prep (minimum input RNA 10 pg, or single cell)
- Low-input cDNA synthesis (minimum input RNA 10 pg, or single cell)
- Degraded low-input RNA-seq library prep (minimum input RNA 10 ng)
- Degraded low-input cDNA synthesis
- Small RNA seq library prep (minimum input RNA 1 ug)
- Low-input small RNA-seq library prep (minimum input RNA 100 ng)
- Single-cell RNA-seq (up to 96 samples)
A QC bioanalyzer run will be added (usually three runs with up to 11 samples per chip).
Quantitative Real-Time PCR is used to very precisely measure the amount of gene expression in cells or tissue of interest, relative to an endogenous control gene such as 18s, b-actin, etc. The two major methods to perform QRTPCR that are supported in the Genome Sciences facility are dual-labeled probe chemistry (TAQMAN) and SYBR green chemistry.
The facility’s recommendation for ease of use is to utilize the pre-optimized Primer/Probe Applied Biosystems Gene Expression Assays (Assays-on-Demand). Penn State has negotiated a good University-wide price, so GEAs should be ordered through ShopOnLion. Taqman Gene Expression Master Mix may be purchased through Genome Sciences iLab under Misc/Order. Also, 384-well barcoded plates and seals may be purchased in the Supply Center or through Genome Sciences iLab under Misc/Order.
The facility can assist in finding other sources should the desired gene not be available at ABI, either with another company or with design assistance.
QuantStudio 12KFlex with 384-well and available OpenArray block is in C2705.
384-well barcoded plates and seals may be purchased in the Supply Center.
Free cloud-based analysis is available through ThermoFisher Digital Science. Upon login, there is a button on the right that says view all applications. Users should click that link and scroll down to the qPCR section.
For those using an unavailable species, or a gene not yet listed, the facility is available to assist with Primer Design and Purchase.
A frequently updated list of validated primer/probe sets for quantitation of a variety of mRNAs is maintained at RealtimePrimers.com.
Investigators who wish to design their own primer/probe sets can use Primer Express, which is available in the facility (Room C2705).
Flanking 5′ (forward) and 3′ (reverse) primers can be made in the Macromolecular Synthesis Core or through external companies such as IDTDNA, Applied Biosystems, etc.
A good source of information describing techniques and including troubleshooting may be found in the 2006 GenomeTech Real-Time PCR Tech Guide, which can be found in OneDrive (Penn State Access ID login required).
Taqman Dual-Labeled Probe (5′-nuclease)
This is an oligo that is complementary to a portion of the gene of interest sequence between PCR primers. This oligo has an fluor attached to the 5′ end, and a quencher attached to the 3′ end. When the oligo has hybridized to the gene sequence and the polymerase incorporates it into the new product, the fluor is allowed to distance itself from the quencher. The rate at which new copies of the gene of interest are generated is inferred by the rate at which the intensity of this free fluor increases.
Dual-labeled probes are more specific than SYBR green because dual-labeled probe chemistry requires specific amplification of the gene of interest in order for fluorescence to be generated. SYBR Green, however, will generate fluorescent signal in the cases of mispriming and the formation of primer dimers. In addition, two different genes of interest may be amplified in one reaction and detected independently from one another by using different fluors on each dual-labeled probe. The disadvantage of using dual-labeled probes is that one dual-labeled probe must be purchased for each gene of interest.
SYBR green
This is a dye that will bind to double-stranded nucleic acid. As primers anneal and the polymerase extends to make new copies of the gene of interest, the amount of SYBR green increases proportionately. The rate at which new copies of the gene of interest are generated is inferred by the rate at which SYBR green fluorescence increases.
qPCR Details
For details on any qPCR information, call 717-531-5823 or email Genome Sciences at genomesciences@pennstatehealth.psu.edu.
Penn State College of Medicine has partnered with Genewiz for Sanger DNA sequencing services.
See Genewiz Sanger submission guidelines here.
Ordering Instructions
- Register for a Genewiz account with the investigator’s lab’s specific Genewiz account ID.
- Once logged in to Genewiz, select “Create a Sequencing Order” from the upper left quadrant of the account home page.
- Select the sequencing order options that best meet project requirements.
- Enter all order information into the online order form or Excel order form.
- Once the Sanger DNA sequencing order is complete, print the order receipt, affix labeled samples to order receipt and place into a Genewiz bubble bag for sample submission.
- Place the Genewiz bubble bag containing the order receipt and samples in the Genewiz Drop Box at Penn State College of Medicine in Room C2705 by 3 p.m. weekdays. (Complete the sample log when dropping off samples at each Genewiz Drop Box to ensure effective submission.)
- Sequencing results will be accessible within the Genewiz account by 5 p.m. the business day following sample submission.
Genewiz Technical Support
Email: dnaseq@genewiz.com
Phone: 908-222-0711, ext. 2
Pricing inquiries: Contact Xuan Pan, Manager, Global Business and Account Development, xuan.pan@genewiz.com or 908-222-0711, ext. 3555
Other DNA Sequencing Services
For additional questions regarding DNA sequencing, contact Sirisha Pochareddy at spochareddy@pennstatehealth.psu.edu.
The Genome Sciences facility uses the Illumina microarray technology Illumina platform for DNA methylation and genotyping arrays.
Genotyping arrays: For focused genotyping/CNV/Methylation studies, the Infinium genotyping arrays offer an exceptional coverage of valuable genomic regions at an affordable price. Investigators to choose from a wide variety of Illumina arrays for specific research aims.
Methylation arrays: Infinium methylation technology allows quantitative measurement of cytosine methylation at the level of single CpG sites, at a genome-wide scale.
Investigators will receive a GenomeStudio project for methylation and other genotyping projects.
The Department of Public Health Sciences is also available for assistance in bioinformatics related to microarray analysis.
Ingenuity Pathway Analysis software is available for the analysis of genomic, transcriptomic and proteomic data.
To obtain an IPA login with the University’s license, please call the Penn State Health Service Desk at 833-577-HELP (4357) and request access to the Ingenuity Pathway Analysis license.
Currently, access is free, and the software may be accessed from any location with an internet connection.
shRNA Plasmid Library clone requests are available through iLab by choosing “OTHER Services” and “Non-Sequencing Request.”
Investigators should follow the instructions in iLab so that the request form loads correctly.
Information on the libraries that will be needed at time of request is located on the network drive in the \smb.files.pennstatehealth.net\cores\ResultsDropoffPickup\GenomeSciences\Incoming\shRNA-List subfolder.
The clones are all catalogued at the Broad Institute. Investigators can search for additional information at the Broad portal here.
Procedures, Protocols and Forms
All RNA (total or mRNA/poly-A+) must be assessed for quality before use in array analysis. It is also recommended prior to any applications, as good-quality RNA is the foundation of all subsequent work and helps to ensure quality and validity to future experiments.
At least 3 µl of RNA at a concentration greater than 0.1-0.5 µg/µl must be provided in a well-labeled tube for the quality control analysis.
RNA QC for up to 12 samples can be analyzed on the RNA 6000 Nano chip, and up to 11 samples can be analyzed on the RNA 6000 Pico chip (rare or micro-dissected samples).
These sample images were provided by Darran May, Aubree Hoover and Tom Sims as part of a presentation on the interpretation of Bioanalyzer 2100 RNA analysis.
Image of Perfect (Intact Eukaryotic) Total RNA
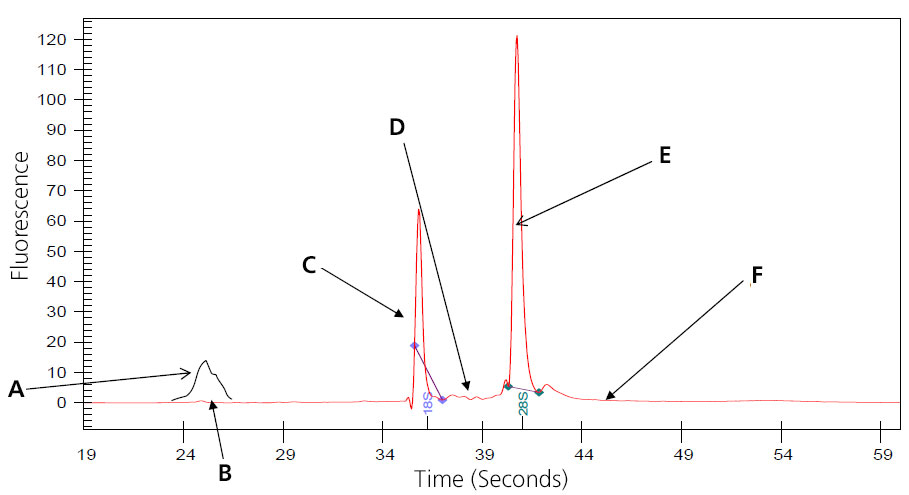
A: 5S subunit; prep-dependent.
B: There maybe a small peak present at approximately 24 seconds that represents 5s, 5.8s and tRNA. This is especially noted with phenol or Trizol extraction, and is eliminated when total RNA is prepped using Qaigen columns, which remove the small RNAs. (Substitute 16S and 23S for prokaryotic samples.)
C: Distinct 18s ribosomal subunit.
D: No small, well-defined peaks between ribosomal peaks.
E: Distinct 28S ribosomal subunit (usually approximately twice 18S).
F: Flat baseline throughout electropherogram.
Samples that result in the above electropherogram image are good to use for message RNA preparation.
Image of Partially Digested Total RNA
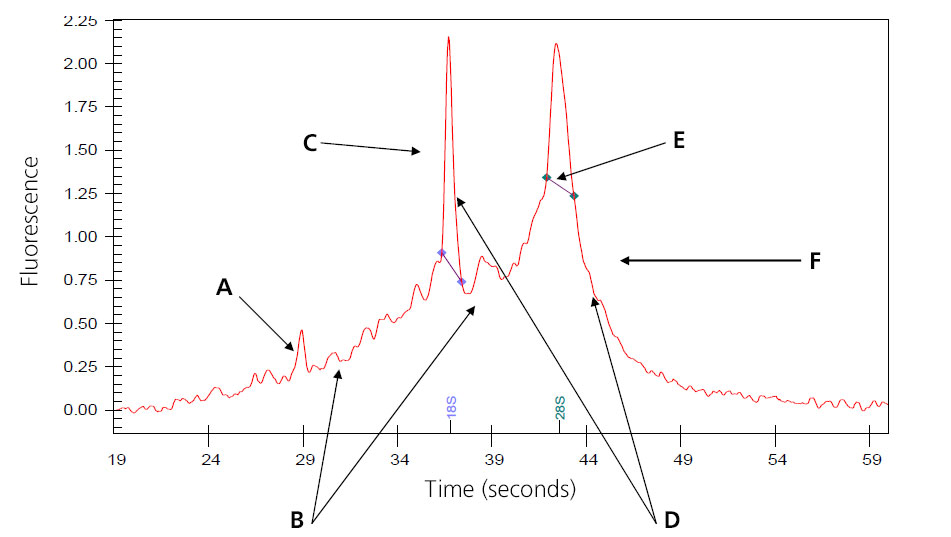
A: Intensities of the smaller degraded RNA increases.
B: Baseline between and to the left of the ribosomal peaks becomes jagged.
C: 18s ribosomal subunit.
D: Intensities of the peaks decrease.
E: 28S ribosomal subunit; in general, the 28S peak begins to degrade first.
F: Peak begins to shift toward the left of the electropherogram.
Samples that result in the above electropherogram image are borderline for message RNA preparation and should be under serious consideration of re-extraction.
Image of Severely Degraded Total RNA
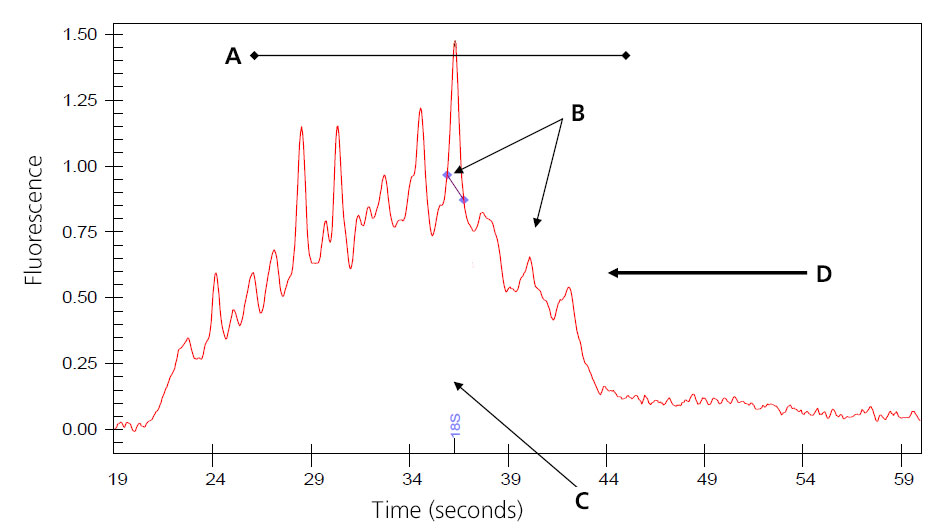
A: Overall decrease in fluorescent signal.
B: Ribosomal peaks mostly degraded; generally 28S peak degrades first.
C: Baseline shifts from flat linke to distinct fragmented peaks.
D: Major shift of products toward left of electropherogram as smaller fragments are created by degradation.
Samples that result in the above electropherogram image should not be used to prep message RNA.
Image of Genomic-DNA-Contaminated Total RNA
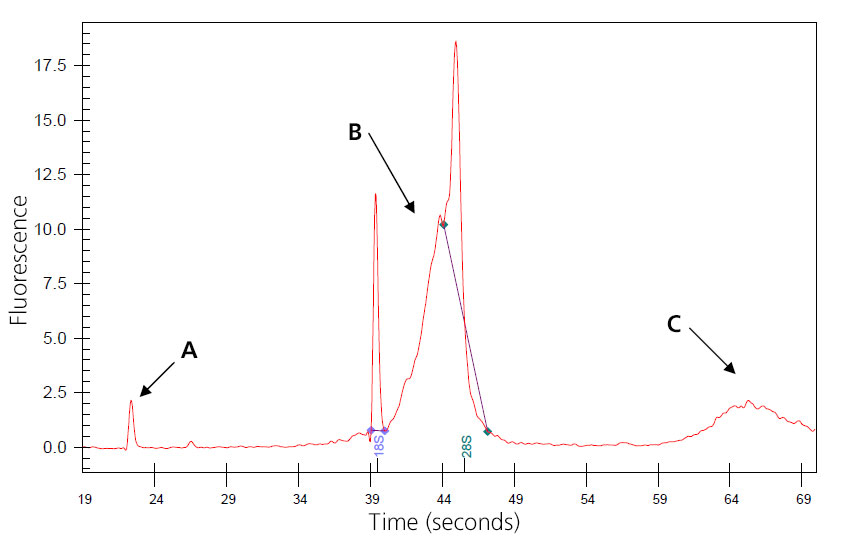
A: Nano peak.
B: Genomic DNA skewing 8S peak.
C: Additional genomic DNA peak.
The sample may wrongly be considered degraded, and concentration may be inflated. Genomic DNA may interfere with downstream reactions if not removed. Treatment can be done during isolation with QIagen RNase-Free DNase or by post-isolation digestion.
(This sample was provided by Joan Tupper.)
For each sample, perform the following, then note the sample number and the nanodrop/bioanalyzer result (ng/µl).
- Add 1 ml Tri-Rgt to tissue; homogenize on ice. It is best to add 500 µl, homogenize, add the remaining 500 µl, and vortex well.
- Add 100 µl BCP; shake for 15 seconds. Let sit at room temperature for 15 minutes. Centrifuge for 16 minutes at maximum (10 to 13K) at 4 degrees C. Pipette top supernatant into new tube.
- Add 500 µl isopropanol; mix well. Let sit for 10 minutes at room temperature. Centrifuge for ine to 12 minutes at maximum (10 to 13K) at 4 to 25 degrees C. Pipette or dump liquid, leaving pellet.
- Add 1 ml 75 percent EtOH and vortex. Centrifuge six minutes at maximum (10 to 13K) at 4 to 25 degrees C. Dump EtOH, leaving pellet; air-dry for approximately eight minutes.
- Dissolve pellet in Rnase-free water (approximately 50 µl or less). Place on ice.
- Test by gel or bioanalyzer to ensure quality of RNA.
- RNA Primer: Sample Prep for Real-Time RT-PCR, a presentation by Eric Lader, PhD, of Qiagen, can be found in OneDrive (Penn State Access ID login required).
- Qiagen’s RNeasy kit is a preferred RNA isolation method and is available in the core supply center.
RNA quality control samples must be submitted by filling out the proper form (RNA Nano, Pico or DNA) and saving it to the Penn State College of Medicine network drive in the Research -> CoreFacility -> Results -> Genome Sciences Incoming -> ~QC Submissions folder.
Note that nano chips hold 12 samples, while pico chips hold 11.
DNA samples are submitted following the same guidelines.
Samples are to be put in the freezer in C2705A in the Genome Sciences QC (Bioanalyzer) box. The freezer is marked with a Genome Sciences Dropoff sign.
Investigators should use their name or other unique description and should ensure the tubes are labeled with something descriptive as well.
Completed results of all work are put in individual researcher folders inside the Research -> CoreFacility -> Results -> Genome Sciences folder.
All publications, press releases or other documents that result from the utilization of any Penn State College of Medicine Institutional Research Resources including funding, tools, services or support are required to credit the core facility and associated RRID for each core used. Use of an instruments in the genome sciences cores should include the following:
“The Genome Sciences Core (RRID:SCR_021123) services and instruments used in this project were funded, in part, by the Pennsylvania State University College of Medicine via the Office of the Vice Dean of Research and Graduate Students and the Pennsylvania Department of Health using Tobacco Settlement Funds (CURE). The content is solely the responsibility of the authors and does not necessarily represent the official views of the University or College of Medicine. The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations or conclusions.”
Failure to do so might result in your access to services being impacted.
Work With Genome Sciences
For information on fees, open an iLab request. The lab can then assist in adding what is needed for proper project planning.
For general inquiries, email GenomeSciences@pennstatehealth.psu.edu.
